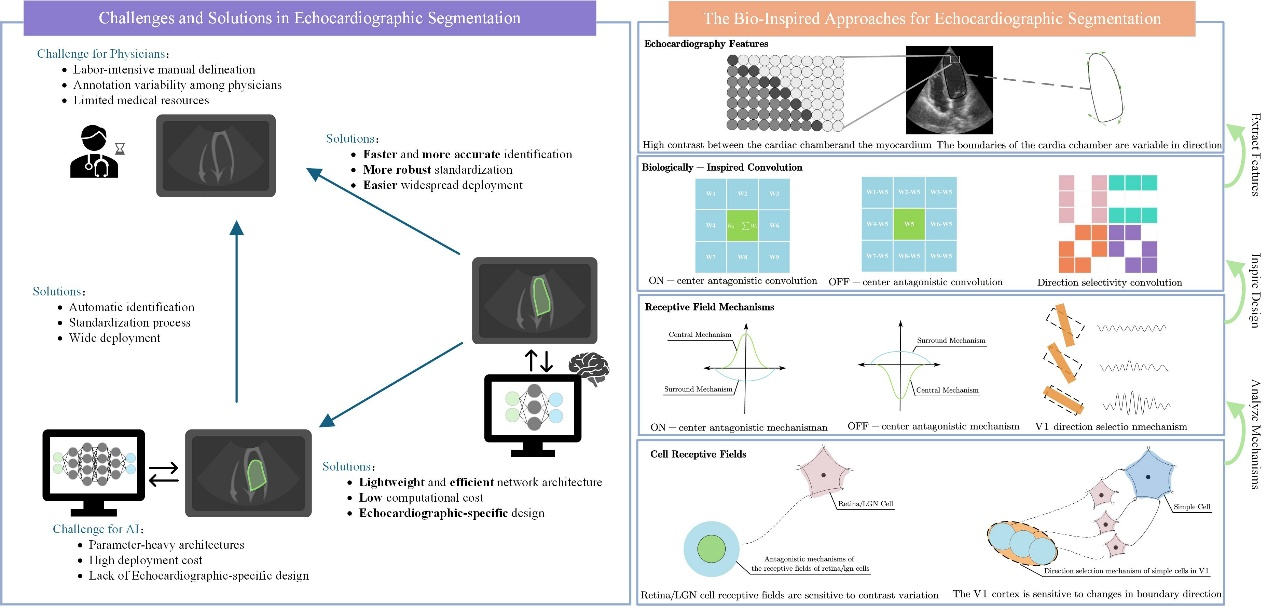
BLENet: a Bio-inspired Lightweight and Efficient Network for Left Ventricle Segmentation in Echocardiography
Project Leaders
Xintao Pan
Project Leaders
Xintao Pan

Project Example
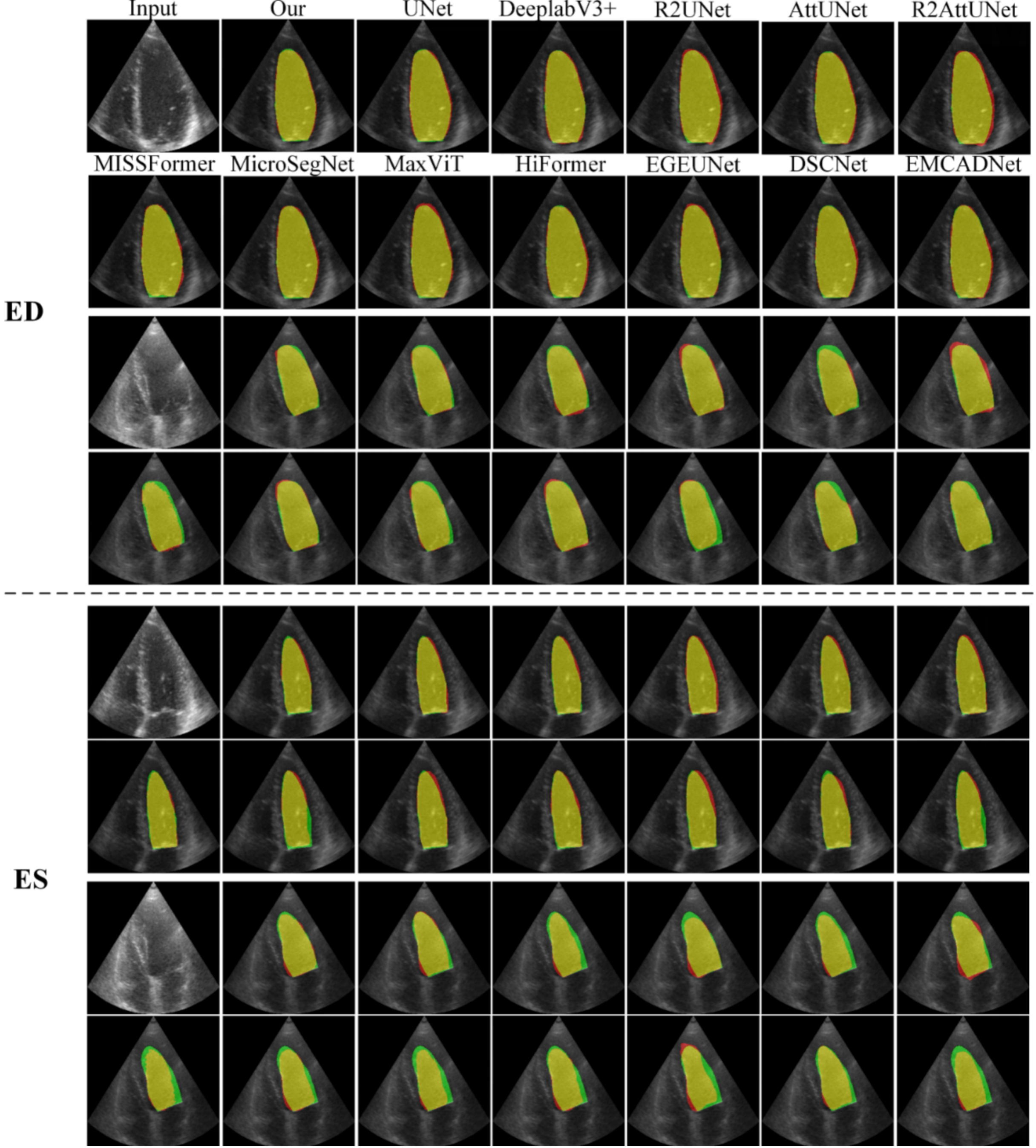
Visualization Comparison of Model Segmentation with SOTA Models on the CAMUS Dataset